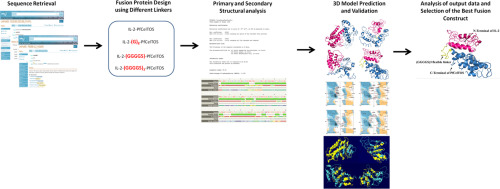
Background
Recombinant protein technology has revolutionized the world of biology and medicine. Following this progress, fusion protein technology, as a novel innovation, has opened new horizons for the development of proteins that do not naturally exist. Fusion proteins are generated via genetically fusing two or more genes coding for separate proteins, thus the product is a single protein having functional properties of both proteins. As an indispensable element in fusion protein construction, linkers are used to separate the functional domains in order to improve their expression, folding and stability.
Method
We computationally fused an antigen and an adjuvant together using different linkers to obtain a two-domain fusion construct which can potentially act as an oral vaccine candidate against malaria. We then predicted the structures computationally to find out the probable folding of each domain in the designed construct.
Results
One of the fusion constructs was selected based on the highest value for C-score. Ramchandran Plot analysis represented that most residues were fallen in favorable regions.
Conclusion
Our in silico analysis showed that (GGGGS)3 linker confers the best structure and stability for our target fusion protein.